Get Eoulsan 2.6.1
 Download Eoulsan Application
Download Eoulsan Application

Welcome to Eoulsan
We developed Eoulsan, a modular and scalable workflow engine dedicated to High Throughput Sequencing analyses. This tool is opinionated as it is focused on production and designed for bioinformaticians.
- Principles
The experimental design of an analysis performed by Eoulsan is stored in a text file, while its workflow steps and parameters are described in a XML file.
These two files ensure flexibility and traceability of the analyses. This approach allows to swiftly resume large analyses upon trouble-shooting, and guarantees reproducibility.
We developed Eoulsan for RNA-Seq, Nanopore RNA-Seq, ChIP-Seq and scRNA-seq workflows.
- Handled infrastructures
Eoulsan can work on a large set of infrastructures (personal workstation, server, cluster and Hadoop cluster). Eoulsan supports most of the job scheduling software (HT-Condor, TGCC, TORQUE, SLURM) to distribute user’s computations.
Moreover the installation of Eoulsan is very easy as users just have to untar its latest archive. The only requirements of Eoulsan are a GNU/Linux distribution and Java.
- Extensibility
To ease the development of new features and modules, Eoulsan reuses the syntax of the XML Galaxy tool description files. Simple Galaxy tools can work under Eoulsan without any modification.
To go further in reproducility of the analyses, Eoulsan promotes Docker and Singularity to deploy image software required its modules.
About this website
This website hosts official binaries of Eoulsan and reference documentation as listed below:
- binaries and source tarballs
- How to install Eoulsan
- A quickstart guide
- and all the reference documentation about the lastest version of the software.
However more up to date workflows can be found the GitHub page of Eoulsan.
What's new in Eoulsan 2.6.1?
The Eoulsan 2.6.1 main new enhancements since its last version are:
- Eoulsan now requires Java 11
- Big refactoring, move many parts of Eoulsan (e.g. the object model) outside Eoulsan in a dedicated library name Kenetre
- Minimap 2.24 binary is now bundled in Eoulsan
- Support of Illumina samplesheet v2 (NextSeq 1000/2000)
- Update versions of many dependencies
- Remove unused code and dependencies (e.g. AWS Elastic MapReduce support)
- Add support for accounting groups with HTCondor.
For the full change log, please report to the dedicated page.
RNA-Seq analysis
Eoulsan contains out the box a fully customizable RNA-Seq pipeline that generates many reports about the performed analysis. The following images are samples of the graphics generated by the normalization and differential analysis steps:
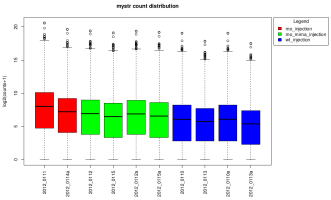
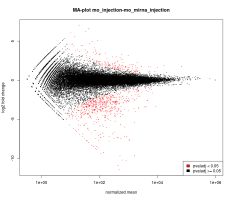
This version currently use DESeq 1 and DESeq 2 for normalization and differential analysis but support for complex designs is only available with DESeq 2.
Object model
Eoulsan contains also a java object model that can be used outside the Eoulsan workflow engine to handle and manipulate NGS data. See the Eoulsan developer wiki for more informations about this part of the project.
Availability
Eoulsan is distributed under the Lesser General Public Licence and CeCill-C.
Funding
This work was supported by the Infrastructures en Biologie Santé et Agronomie (IBiSA), France Genomique.



